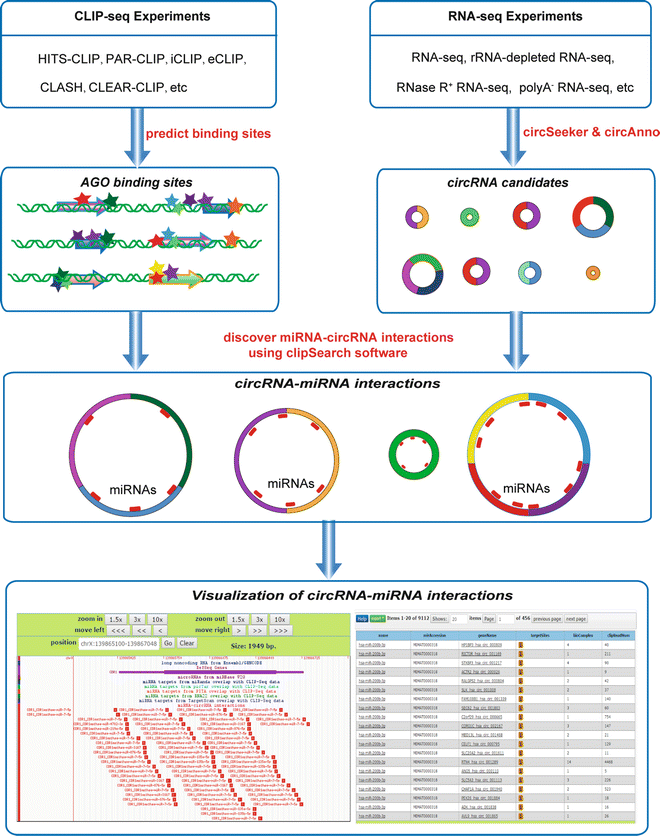
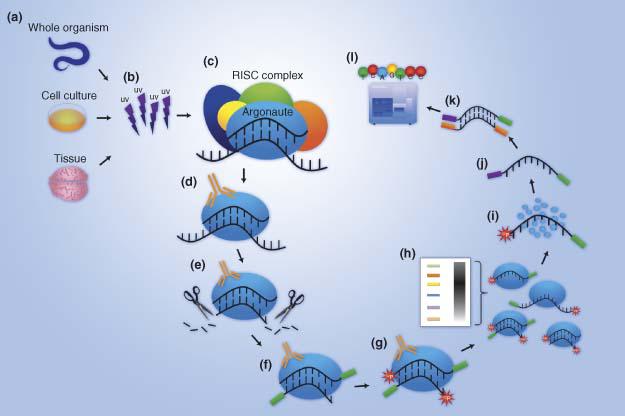
Pipeline for extracting MIWI CLIP-Seq features (see Section 2). The RNA... | Download Scientific Diagram

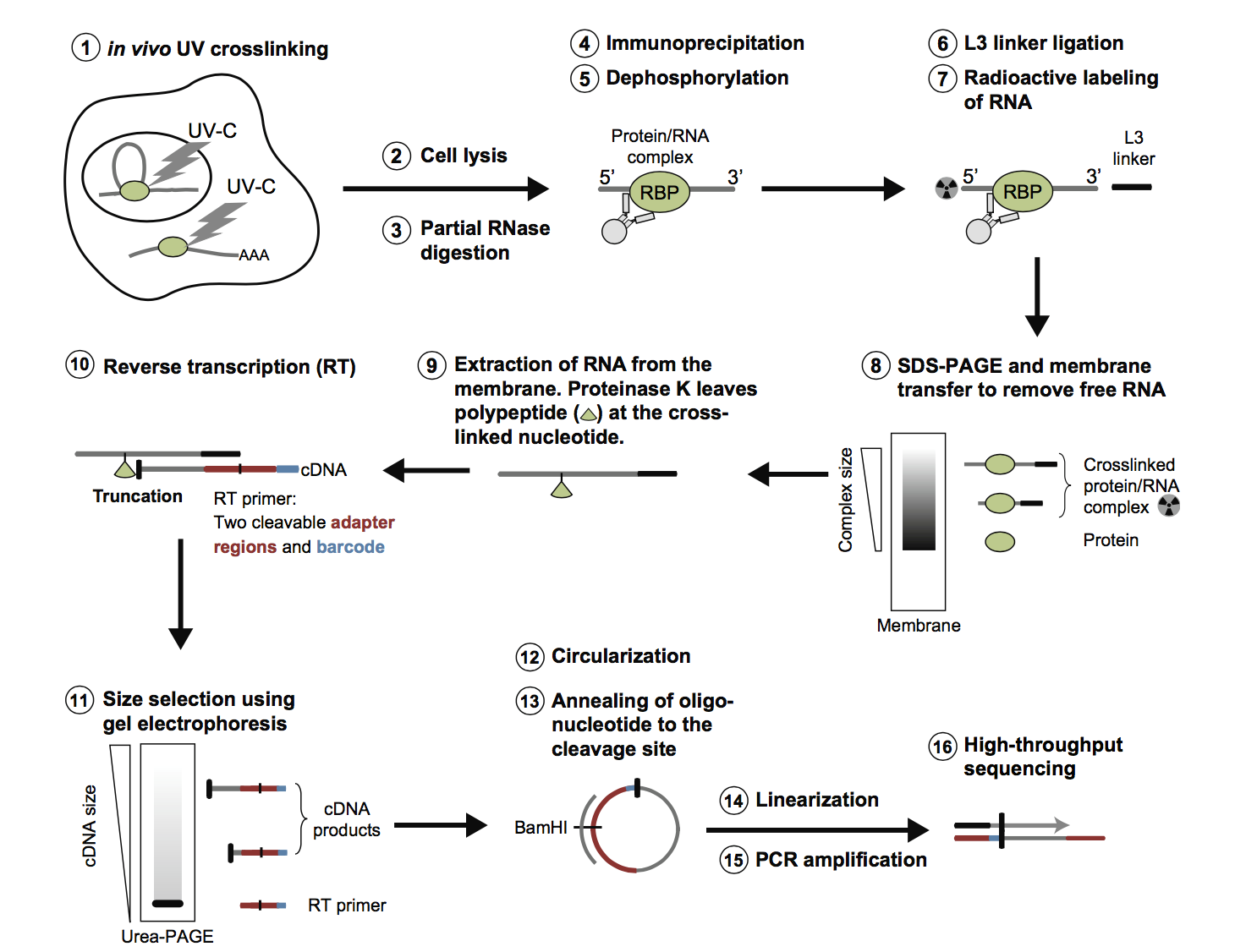
SURF: integrative analysis of a compendium of RNA-seq and CLIP-seq datasets highlights complex governing of alternative transcriptional regulation by RNA-binding proteins | Genome Biology | Full Text

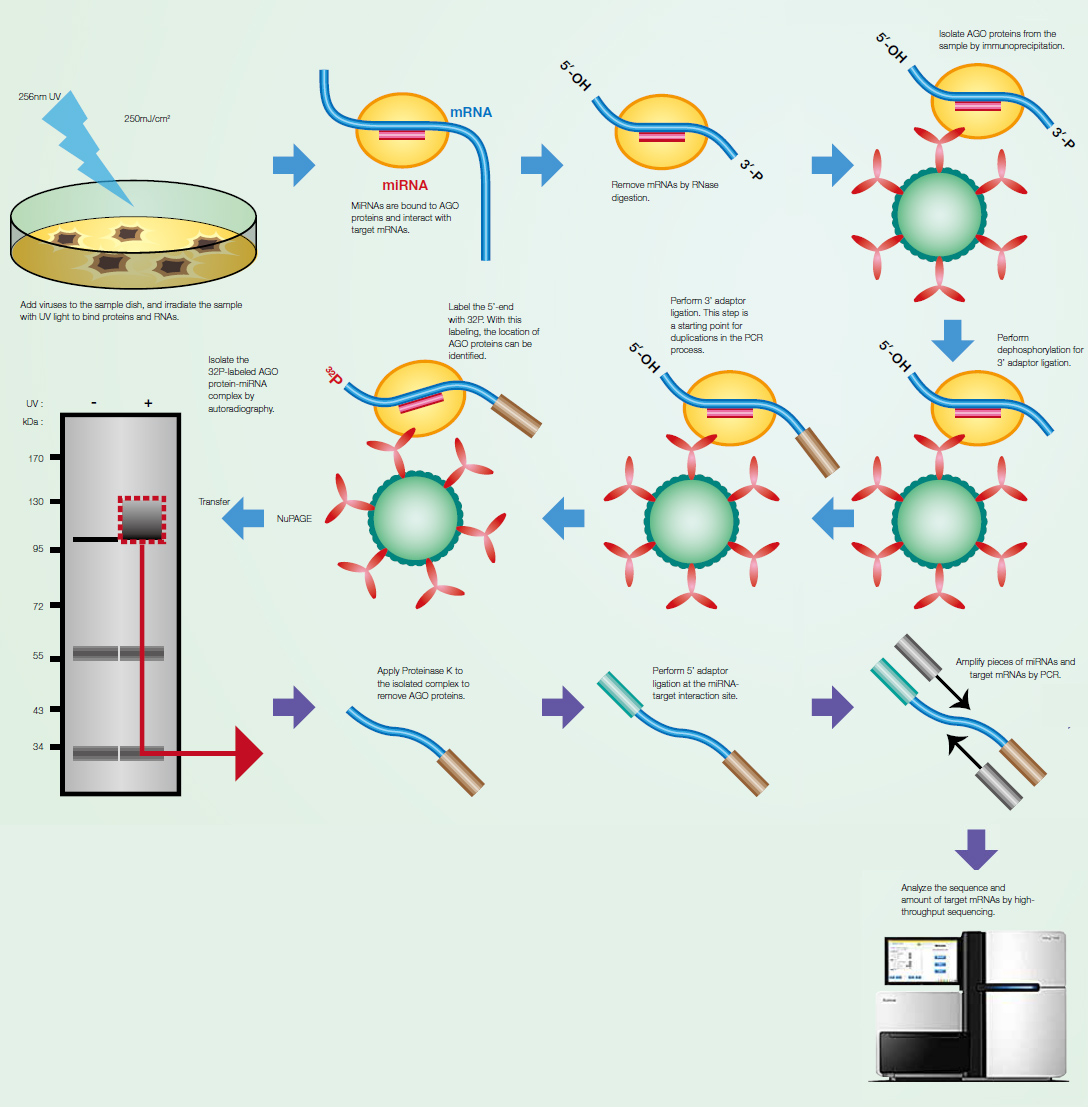
Main steps of the bioinformatics workflow to analyze CLIP-seq data with... | Download Scientific Diagram
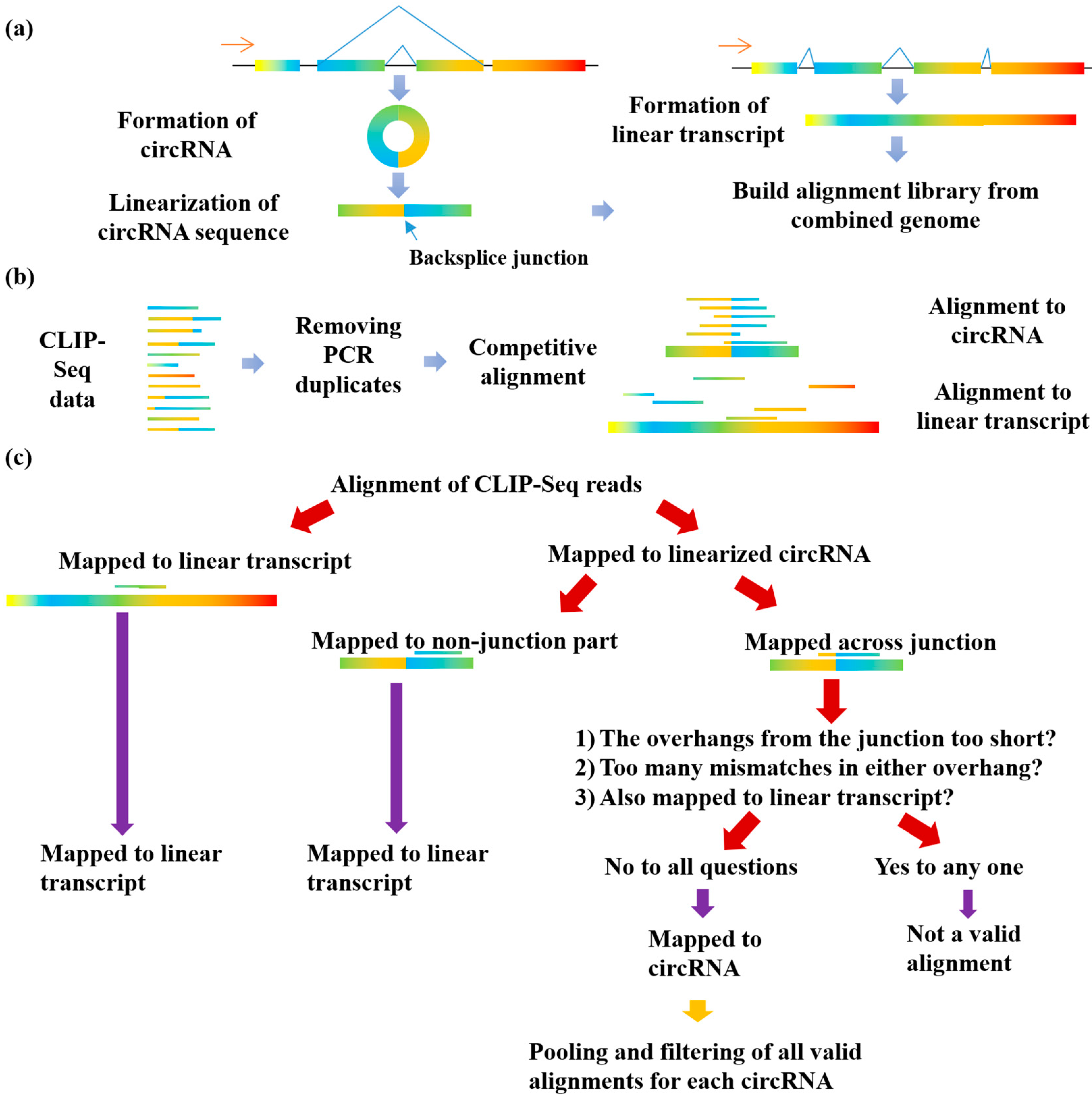
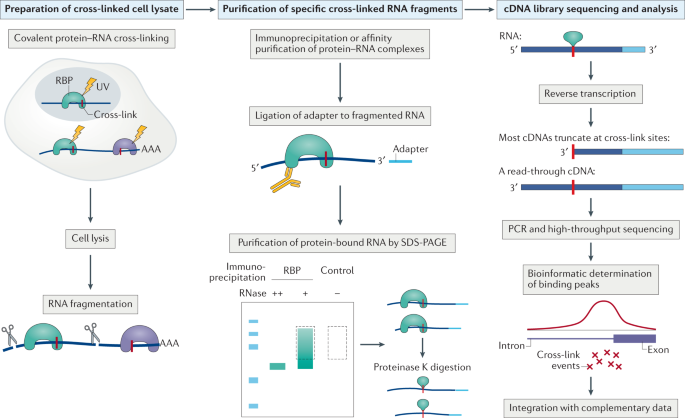
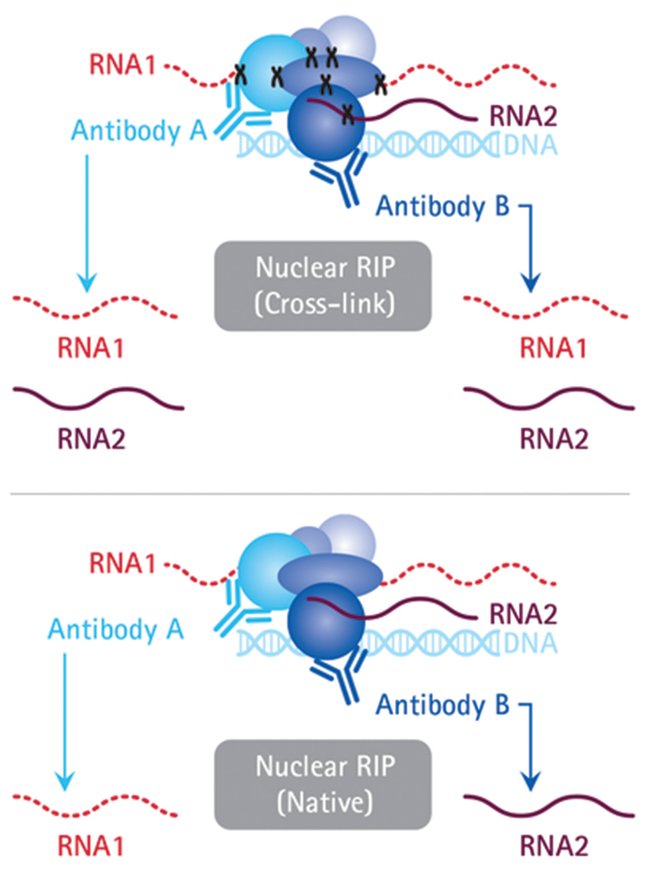
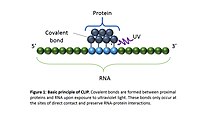
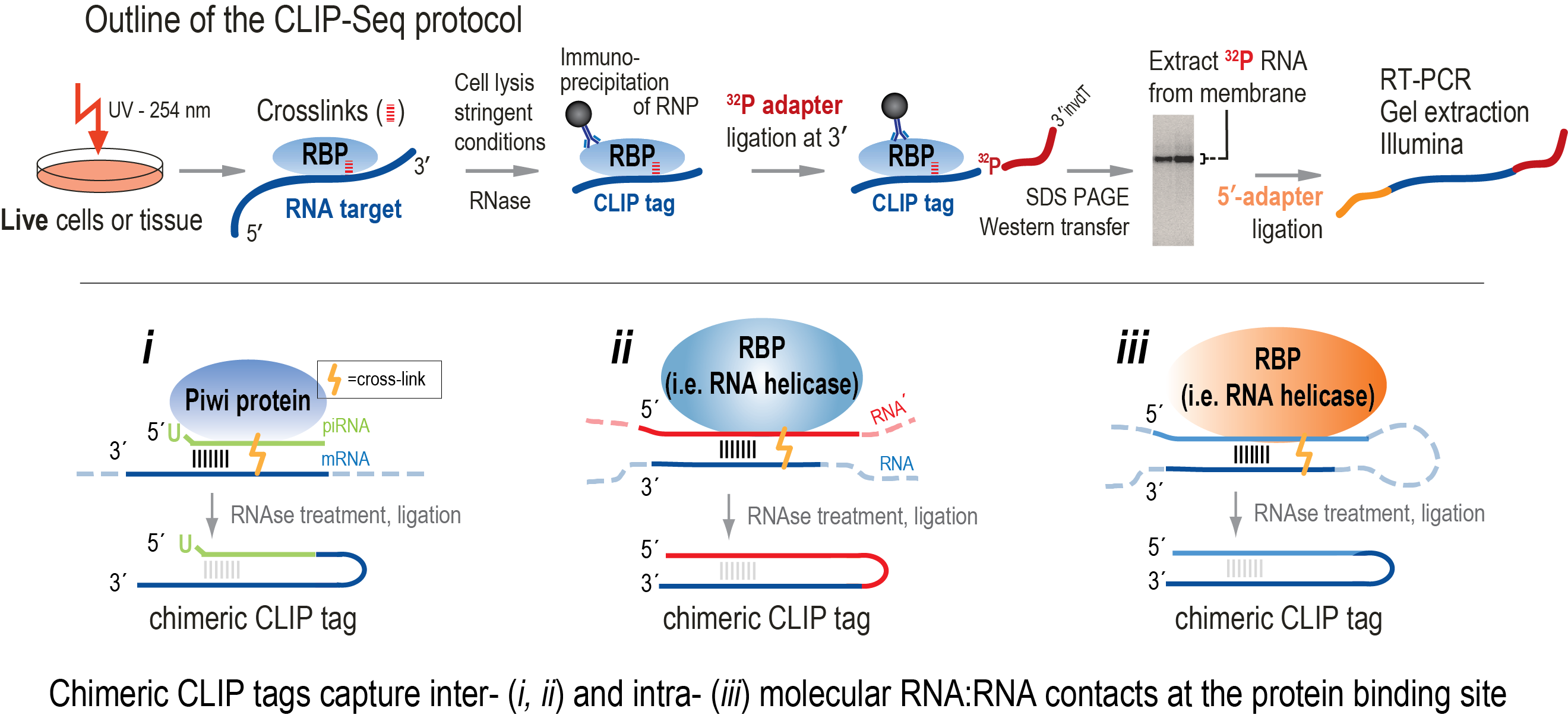

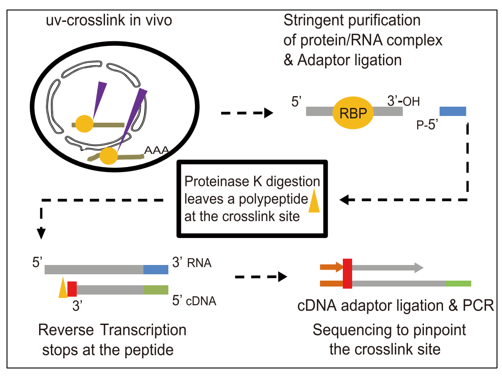
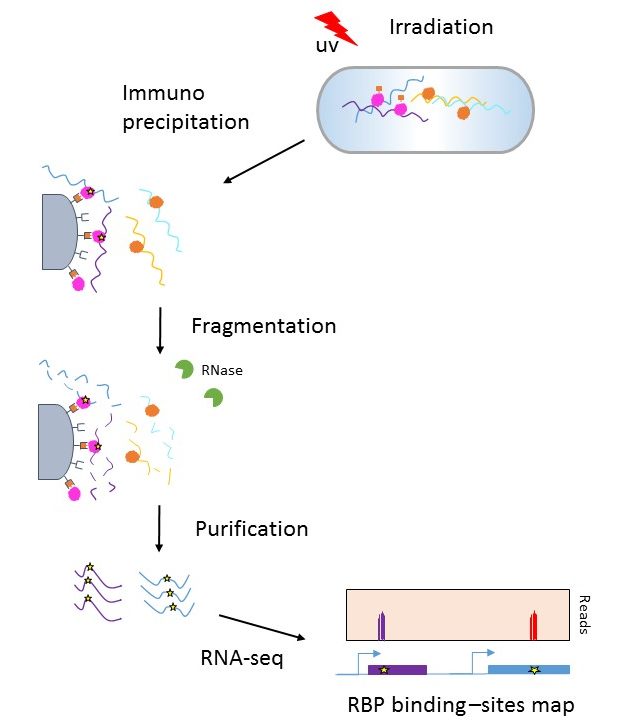


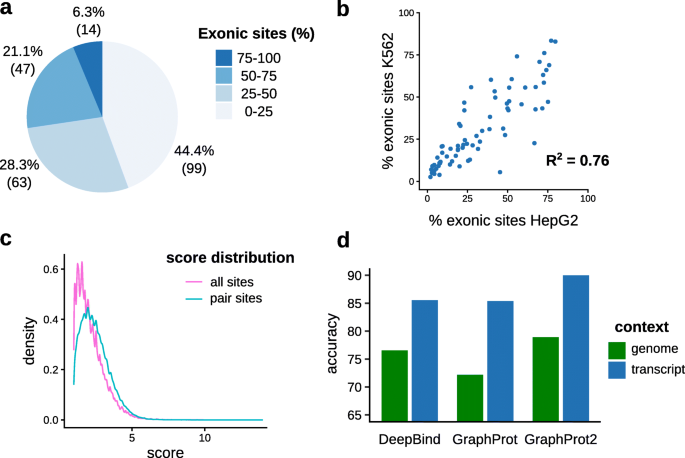
![PDF] Current methods in the analysis of CLIP-Seq data | Semantic Scholar PDF] Current methods in the analysis of CLIP-Seq data | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/e504d75c2c48b19613a796fb54083b71e3781289/2-Figure1-1.png)